

This page gives clear instructions on how to use IPAD-DB. If you have any problems, please do not hesitate to contact Dr. Chong Peng. Email: cpeng@tust.edu.cn.
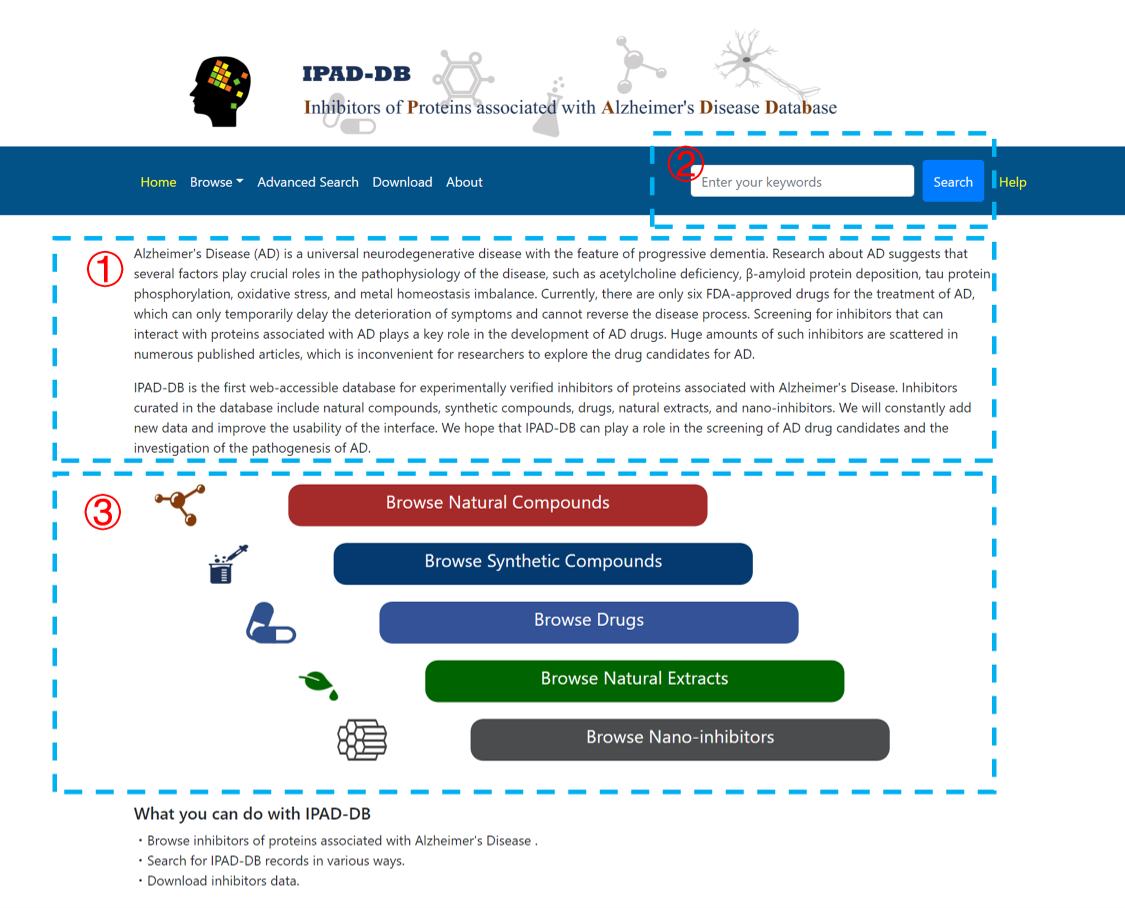
The home page provides a basic introduction to IPAD-DB (1). Users can fill an arbitrary keyword in the search box to make a quick search in the database (2). Users can also browse the data by clicking the categories on the home page (3).
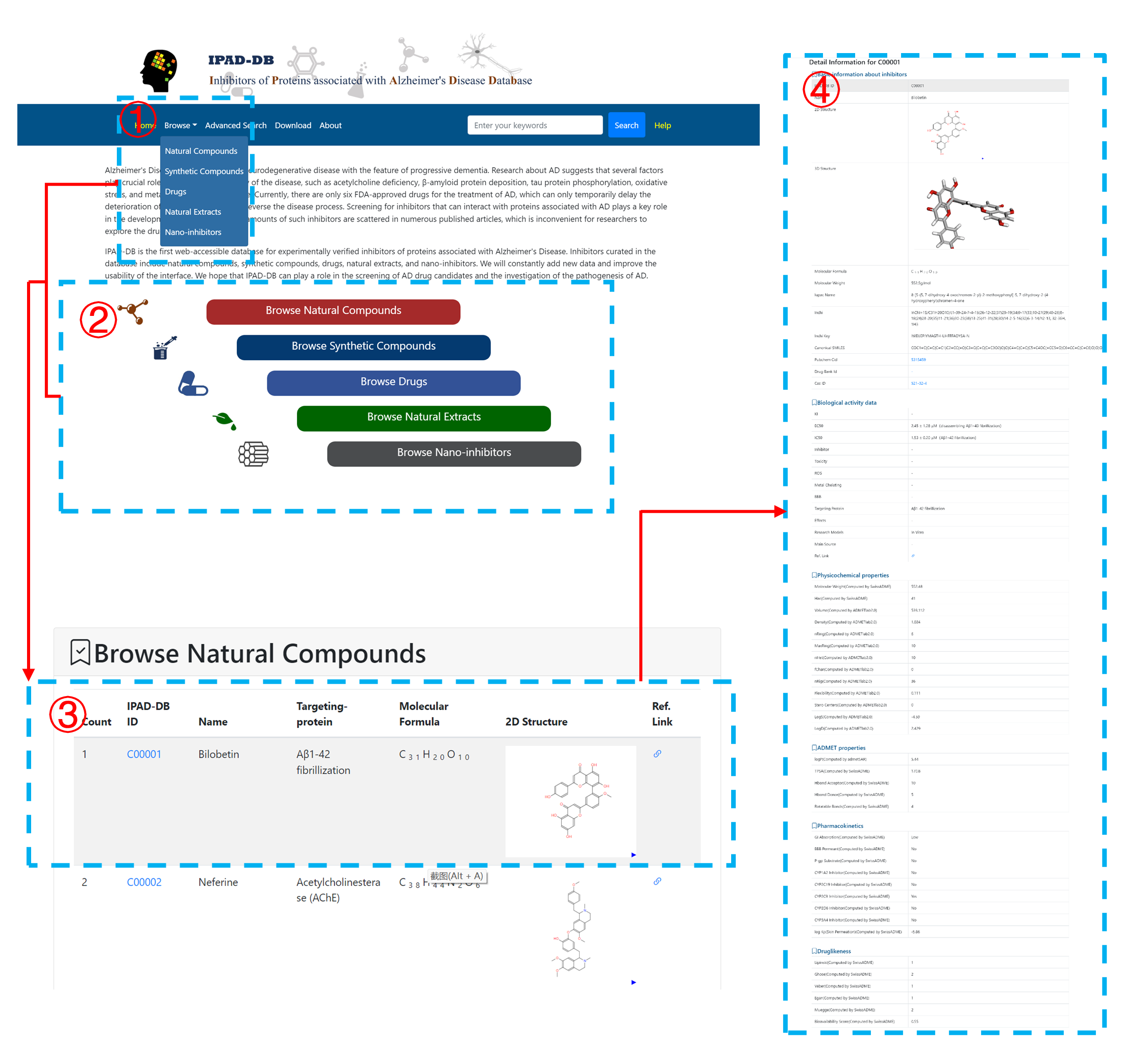
There are five tables on the Browse page. The detailed information can be viewed by clicking the corresponding button (1). Users can also browse the data by clicking the categories on the home page (2). Basic information about inhibitors can be viewed in the list (3). The detailed information can be viewed by clicking the corresponding button (4).
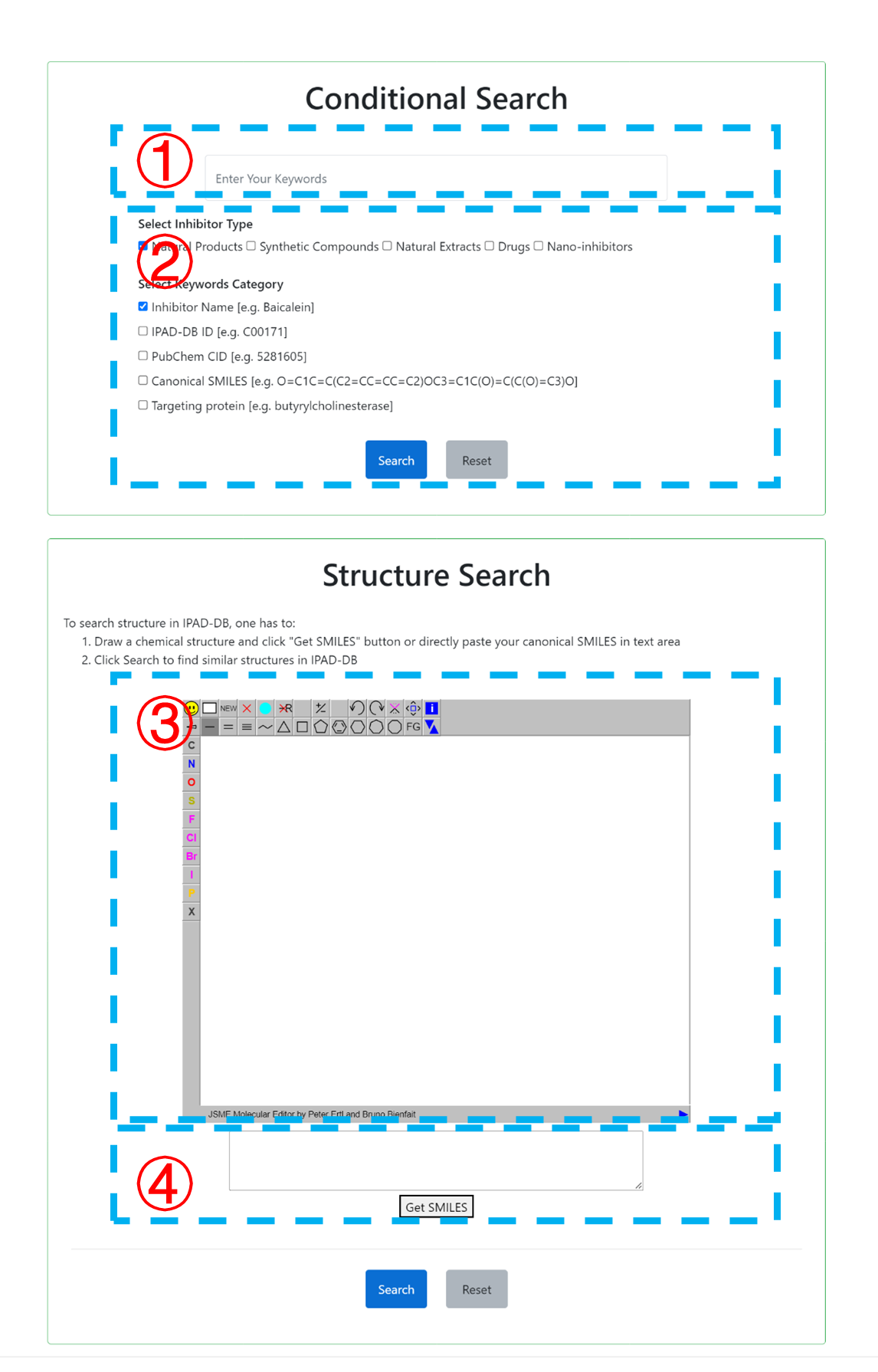
IPAD-DB provides both text-based and structure-based search methods. For text-based search, users can enter the inhibitor name, target protein, molecular SMILES, or IPAD-DB ID in the search box (1). Then, select the inhibitor type and the keywords category (2). For structure-based search, users can draw a query molecule using JSME editor (3), get the SMILES of the molecule (4) and search in the database for compounds with similar structures.
The basic information for five kinds of inhibitors is packaged in csv format. The sdf files of natural compounds, synthetic compounds, and drugs are packaged as compressed files. These files are available and can be freely downloaded.
